Big Data Science with the BD2K-LINCS Data Coordination and Integration Center
WEBRip | English | MP4 + PDF Guides | 960 x 540 | AVC ~67.7 kbps | 29.970 fps
AAC | 128 Kbps | 44.1 KHz | 2 channels | Subs: English (.srt) | 06:14:15 | 505 MB
Genre: eLearning Video / Science, Bioinformatics
WEBRip | English | MP4 + PDF Guides | 960 x 540 | AVC ~67.7 kbps | 29.970 fps
AAC | 128 Kbps | 44.1 KHz | 2 channels | Subs: English (.srt) | 06:14:15 | 505 MB
Genre: eLearning Video / Science, Bioinformatics
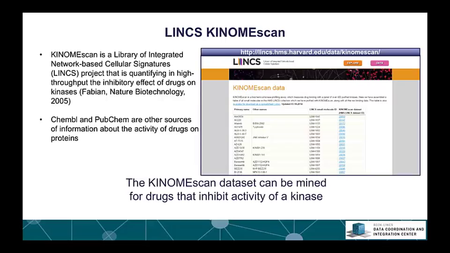
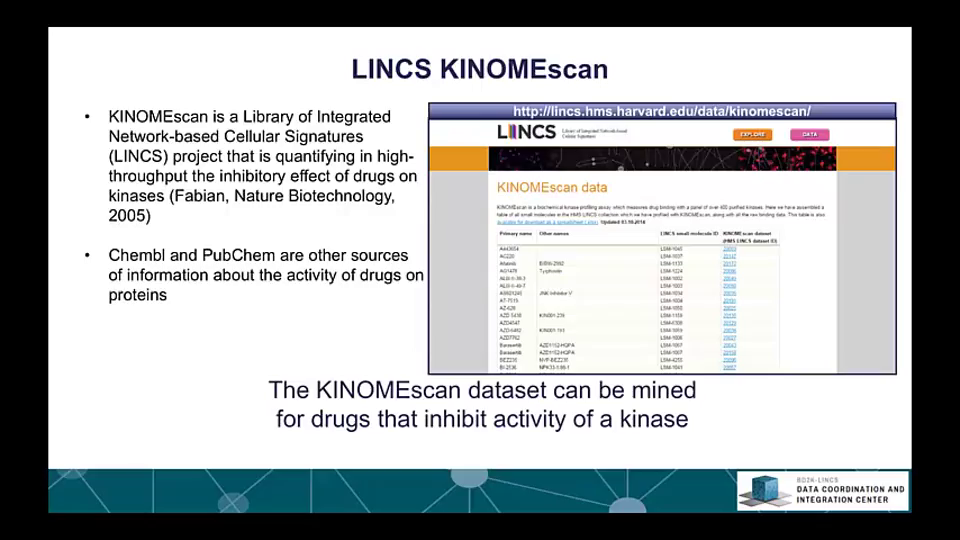
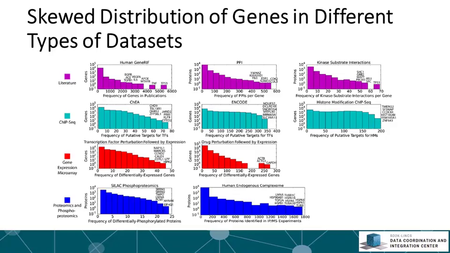
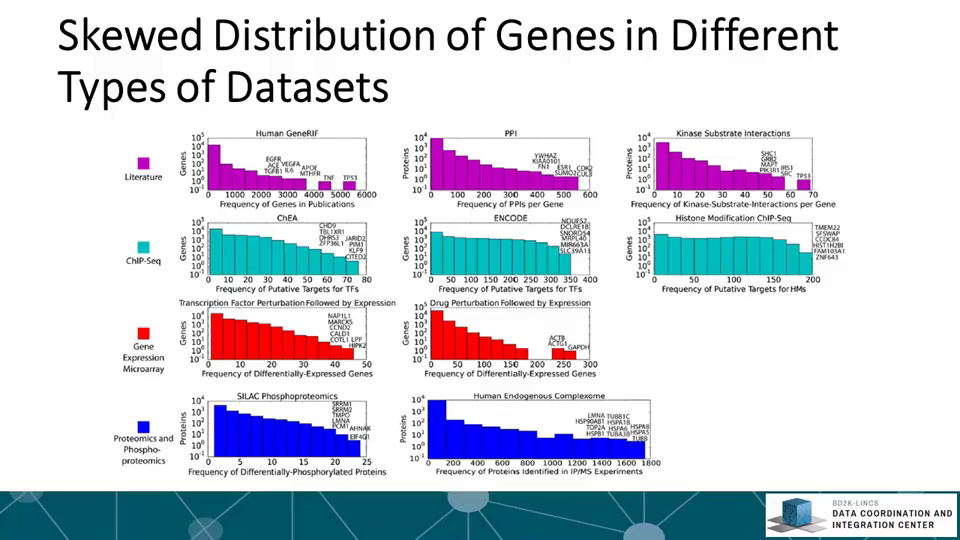
The Library of Integrative Network-based Cellular Signatures (LINCS) is an NIH Common Fund program. The idea is to perturb different types of human cells with many different types of perturbations such as: drugs and other small molecules; genetic manipulations such as knockdown or overexpression of single genes; manipulation of the extracellular microenvironment conditions, for example, growing cells on different surfaces, and more.These perturbations are applied to various types of human cells including induced pluripotent stem cells from patients, differentiated into various lineages such as neurons or cardiomyocytes. Then, to better understand the molecular networks that are affected by these perturbations, changes in level of many different variables are measured including: mRNAs, proteins, and metabolites, as well as cellular phenotypic changes such as changes in cell morphology. The BD2K-LINCS Data Coordination and Integration Center (DCIC) is commissioned to organize, analyze, visualize and integrate this data with other publicly available relevant resources. In this course we briefly introduce the DCIC and the various Centers that collect data for LINCS. We then cover metadata and how metadata is linked to ontologies. We then present data processing and normalization methods to clean and harmonize LINCS data. This follow discussions about how data is served as RESTful APIs. Most importantly, the course covers computational methods including: data clustering, gene-set enrichment analysis, interactive data visualization, and supervised learning. Finally, we introduce crowdsourcing/citizen-science projects where students can work together in teams to extract expression signatures from public databases and then query such collections of signatures against LINCS data for predicting small molecules as potential therapeutics.
Content
Module 1 - The Library of Integrated Network-based Cellular Signatures LINCS Program Overview
Module 2 - Metadata and Ontologies
Module 3 - Serving Data with APIs
Module 4 - Bioinformatics Pipelines
Module 5 - The Harmonizome
Module 6 - Data Normalization
Module 7 - Data_Clustering
Module 8 - Enrichment Analysis
Module 9 - Machine Learning
Module 10 - Benchmarking
Module 11 - Interactive Data Visualization
Module 12 - Crowdsourcing Projects
also You can watch my other last: Coursera-posts
General
Complete name : 02_Processing_Datasets__Part_1_8-27.mp4
Format : MPEG-4
Format profile : Base Media
Codec ID : isom
File size : 12.4 MiB
Duration : 8mn 27s
Overall bit rate : 204 Kbps
Writing application : Lavf55.19.104
Video
ID : 1
Format : AVC
Format/Info : Advanced Video Codec
Format profile : Main@L3.1
Format settings, CABAC : Yes
Format settings, ReFrames : 4 frames
Codec ID : avc1
Codec ID/Info : Advanced Video Coding
Duration : 8mn 27s
Bit rate : 67.7 Kbps
Width : 960 pixels
Height : 540 pixels
Display aspect ratio : 16:9
Original display aspect rat : 16:9
Frame rate mode : Constant
Frame rate : 29.970 fps
Color space : YUV
Chroma subsampling : 4:2:0
Bit depth : 8 bits
Scan type : Progressive
Bits/(Pixel*Frame) : 0.004
Stream size : 4.09 MiB (33%)
Writing library : x264 core 138
Encoding settings : cabac=1 / ref=3 / deblock=1:0:0 / analyse=0x1:0x111 / me=hex / subme=7 / psy=1 / psy_rd=1.00:0.00 / mixed_ref=1 / me_range=16 / chroma_me=1 / trellis=1 / 8x8dct=0 / cqm=0 / deadzone=21,11 / fast_pskip=1 / chroma_qp_offset=-2 / threads=12 / lookahead_threads=2 / sliced_threads=0 / nr=0 / decimate=1 / interlaced=0 / bluray_compat=0 / constrained_intra=0 / bframes=3 / b_pyramid=2 / b_adapt=1 / b_bias=0 / direct=1 / weightb=1 / open_gop=0 / weightp=2 / keyint=250 / keyint_min=25 / scenecut=40 / intra_refresh=0 / rc_lookahead=40 / rc=crf / mbtree=1 / crf=28.0 / qcomp=0.60 / qpmin=0 / qpmax=69 / qpstep=4 / ip_ratio=1.40 / aq=1:1.00
Language : English
Audio
ID : 2
Format : AAC
Format/Info : Advanced Audio Codec
Format profile : LC
Codec ID : 40
Duration : 8mn 27s
Bit rate mode : Constant
Bit rate : 128 Kbps
Channel(s) : 2 channels
Channel positions : Front: L R
Sampling rate : 44.1 KHz
Compression mode : Lossy
Stream size : 7.74 MiB (63%)
Language : English
Complete name : 02_Processing_Datasets__Part_1_8-27.mp4
Format : MPEG-4
Format profile : Base Media
Codec ID : isom
File size : 12.4 MiB
Duration : 8mn 27s
Overall bit rate : 204 Kbps
Writing application : Lavf55.19.104
Video
ID : 1
Format : AVC
Format/Info : Advanced Video Codec
Format profile : Main@L3.1
Format settings, CABAC : Yes
Format settings, ReFrames : 4 frames
Codec ID : avc1
Codec ID/Info : Advanced Video Coding
Duration : 8mn 27s
Bit rate : 67.7 Kbps
Width : 960 pixels
Height : 540 pixels
Display aspect ratio : 16:9
Original display aspect rat : 16:9
Frame rate mode : Constant
Frame rate : 29.970 fps
Color space : YUV
Chroma subsampling : 4:2:0
Bit depth : 8 bits
Scan type : Progressive
Bits/(Pixel*Frame) : 0.004
Stream size : 4.09 MiB (33%)
Writing library : x264 core 138
Encoding settings : cabac=1 / ref=3 / deblock=1:0:0 / analyse=0x1:0x111 / me=hex / subme=7 / psy=1 / psy_rd=1.00:0.00 / mixed_ref=1 / me_range=16 / chroma_me=1 / trellis=1 / 8x8dct=0 / cqm=0 / deadzone=21,11 / fast_pskip=1 / chroma_qp_offset=-2 / threads=12 / lookahead_threads=2 / sliced_threads=0 / nr=0 / decimate=1 / interlaced=0 / bluray_compat=0 / constrained_intra=0 / bframes=3 / b_pyramid=2 / b_adapt=1 / b_bias=0 / direct=1 / weightb=1 / open_gop=0 / weightp=2 / keyint=250 / keyint_min=25 / scenecut=40 / intra_refresh=0 / rc_lookahead=40 / rc=crf / mbtree=1 / crf=28.0 / qcomp=0.60 / qpmin=0 / qpmax=69 / qpstep=4 / ip_ratio=1.40 / aq=1:1.00
Language : English
Audio
ID : 2
Format : AAC
Format/Info : Advanced Audio Codec
Format profile : LC
Codec ID : 40
Duration : 8mn 27s
Bit rate mode : Constant
Bit rate : 128 Kbps
Channel(s) : 2 channels
Channel positions : Front: L R
Sampling rate : 44.1 KHz
Compression mode : Lossy
Stream size : 7.74 MiB (63%)
Language : English
Screenshots
Exclusive eLearning Videos ParRus-blog ← add to bookmarks