Schrodinger Suites 2021-3 | 6.5 Gb
The software developer Schrödinger continuously strive to develop scientific solutions that push the boundaries of molecular design and are delighted to share enhanced drug discovery and materials workflows in Schrödinger’s 2021-3 software release.
Schrödinger Software Release 2021-3
This quarterly release includes:
- Intuitive enhancement of the protein preparation workflow and kinase conservation annotations for structure enablement
- Addition of filters to improve drug-likeness for medical chemistry design using Ligand Designer
- A new diversity approach to select compounds for Active Learning Glide workflows for hit discovery and lead optimization
- Greater control of custom R-group enumerations for hit discovery and lead optimization for multiple simultaneous substitutions
Workflows & Pipelining [KNIME Extensions]
Maestro Graphical Interface
- Recognize nucleic acid macromolecules as receptors [2021-3]
- Improved organization of multi-residue ligands (e.g. peptides) in the Structure Hierarchy [2021-3]
- New preference to “Allow nucleic acid molecules as ligands” [2021-3]
- New preference to “Avoid custom dialogs” (Windows 10 only) [2021-3]
- Label “Residue Information” on a selected atom [2021-3]
- New Protein Preparation Workflow as the default for Favorites toolbar and Prepare banner [2021-3]
Force Field
- Improved accuracy of intracyclic torsion parameters for sulfonyl pyrroles [2021-3]
Workflows & Pipelining [KNIME Extensions]
KNIME in LiveDesign:
Upload as LiveDesign model node [2021-3]
- The extra argument and extra file fields are populated automatically
- The model folder can be set in the Preferences
Model administration [2021-3]
- New node to list the deployed protocols and models
- Models can be archived using LiveDesign_admin.py
Structure Enablement
Protein Preparation
- First full release out of beta [2021-3]
- Renumber antibody residues using standard residue numbering for antibody naming scheme selected [2021-3]
Cryo-EM Model Refinement
- First full release out of beta for GlideEM and PHENIX/OPLS3e [2021-3]
- Support PHENIX 1.19.2 [2021-3]
Multiple Sequence Viewer/Editor
- Added kinase conservation annotation [2021-3]
- Homology modeling workflow improved to simplify template inclusion via file import or homolog searching [2021-3]
Binding Site & Structure Analysis
Desmond Molecular Dynamics
- Added support for Parrinello-Rahman barostat that allows for unit cell angle changes [2021-3]
- Added lipid-specific trajectory analysis [2021-3]
WaterMap
- Default forcefield for command line invocation changed to OPLS4 [2021-3]
Medicinal Chemistry Design
Ligand Designer
- Import custom R-groups for ideation directly from Ligand Designer [2021-3]
- Improve druglikeness of ideas generated by reaction-based enumeration by filtering out PAINS compounds and those that violate REOS or Schrodinger’s standard filters [2021-3]
Conformational Analysis
Quantum Mechanics
- Added wB97X-D3 and wB97We X-V functional [2021-3]
- Improve control over the maximum number of simultaneous subjobs and parallelization with OpenMP threads [2021-3]
- Added ethanol, methanol, DMSO, and acetonitrile as solvents when predicting circular dichroism [2021-3]
Property Prediction
FEP+
- Simplify map visualization by clustering nodes with the same ligand in different charge/tautomer/conformation states [2021-3]
Protein FEP
- Examine correlations between experimental and FEP predicted affinities [2021-3]
Quantum Mechanics
- Enable alignment of theoretical and experimental IR spectra [2021-3]
Hit Discovery
Enumeration
- Support global and per-library concentrations when performing multiple R-group enumerations simultaneously [2021-3]
Pharmacophore Modeling
- Significant speedup of in-place pharmacophore screens against hypothesis with large numbers of features where few features are required to be matched. [2021-3]
Ligand Docking
- New input file format for Active Learning Glide simulations [2021-3]
- Allow separate specification of the number of simultaneous docking and ML prediction simulations to control license usage in Active Learning Glide [2021-3]
- Added counts of ligands that failed to dock for various reasons [2021-3]
- Return a CSV file of screened ligands by default [2021-3]
- Support Hydrogen bond and positional constraints in WScore [2021-3]
ABFEP
- Improved accuracy by splitting complex leg lambda schedules for dihedral restraints from distance and angle restraints and ensuring overlap of cross-link restraints with the schedules of charge and vdW [2021-3]
Maestro Graphical Interface
- Recognize nucleic acid macromolecules as receptors [2021-3]
- Improved organization of multi-residue ligands (e.g. peptides) in the Structure Hierarchy [2021-3]
- New preference to “Allow nucleic acid molecules as ligands” [2021-3]
- New preference to “Avoid custom dialogs” (Windows 10 only) [2021-3]
- Label “Residue Information” on a selected atom [2021-3]
- New Protein Preparation Workflow as the default for Favorites toolbar and Prepare banner [2021-3]
Force Field
- Improved accuracy of intracyclic torsion parameters for sulfonyl pyrroles [2021-3]
Workflows & Pipelining [KNIME Extensions]
KNIME in LiveDesign:
Upload as LiveDesign model node [2021-3]
- The extra argument and extra file fields are populated automatically
- The model folder can be set in the Preferences
Model administration [2021-3]
- New node to list the deployed protocols and models
- Models can be archived using LiveDesign_admin.py
Structure Enablement
Protein Preparation
- First full release out of beta [2021-3]
- Renumber antibody residues using standard residue numbering for antibody naming scheme selected [2021-3]
Cryo-EM Model Refinement
- First full release out of beta for GlideEM and PHENIX/OPLS3e [2021-3]
- Support PHENIX 1.19.2 [2021-3]
Multiple Sequence Viewer/Editor
- Added kinase conservation annotation [2021-3]
- Homology modeling workflow improved to simplify template inclusion via file import or homolog searching [2021-3]
Binding Site & Structure Analysis
Desmond Molecular Dynamics
- Added support for Parrinello-Rahman barostat that allows for unit cell angle changes [2021-3]
- Added lipid-specific trajectory analysis [2021-3]
WaterMap
- Default forcefield for command line invocation changed to OPLS4 [2021-3]
Medicinal Chemistry Design
Ligand Designer
- Import custom R-groups for ideation directly from Ligand Designer [2021-3]
- Improve druglikeness of ideas generated by reaction-based enumeration by filtering out PAINS compounds and those that violate REOS or Schrodinger’s standard filters [2021-3]
Conformational Analysis
Quantum Mechanics
- Added wB97X-D3 and wB97We X-V functional [2021-3]
- Improve control over the maximum number of simultaneous subjobs and parallelization with OpenMP threads [2021-3]
- Added ethanol, methanol, DMSO, and acetonitrile as solvents when predicting circular dichroism [2021-3]
Property Prediction
FEP+
- Simplify map visualization by clustering nodes with the same ligand in different charge/tautomer/conformation states [2021-3]
Protein FEP
- Examine correlations between experimental and FEP predicted affinities [2021-3]
Quantum Mechanics
- Enable alignment of theoretical and experimental IR spectra [2021-3]
Hit Discovery
Enumeration
- Support global and per-library concentrations when performing multiple R-group enumerations simultaneously [2021-3]
Pharmacophore Modeling
- Significant speedup of in-place pharmacophore screens against hypothesis with large numbers of features where few features are required to be matched. [2021-3]
Ligand Docking
- New input file format for Active Learning Glide simulations [2021-3]
- Allow separate specification of the number of simultaneous docking and ML prediction simulations to control license usage in Active Learning Glide [2021-3]
- Added counts of ligands that failed to dock for various reasons [2021-3]
- Return a CSV file of screened ligands by default [2021-3]
- Support Hydrogen bond and positional constraints in WScore [2021-3]
ABFEP
- Improved accuracy by splitting complex leg lambda schedules for dihedral restraints from distance and angle restraints and ensuring overlap of cross-link restraints with the schedules of charge and vdW [2021-3]
Schrödinger Software provide accurate, reliable, and high performance computational technology to solve real-world problems in life science research. It can be used to build, edit, run and analyse molecules.
The Schrödinger-Suite of applications have a graphical user interface called Maestro. Using the Maestro you can prepare your structure for refinement.
The following products are available: CombiClide, ConfGen, Core Hopping, Desmond, Epik, Glide, Glide, Impact, Jaguar ( high-performance ab initio package), Liaison, LigPrep, MacroModel, MCPRO+, Phase, Prime, PrimeX, QikProp, QSite, Semi-Empirical, SiteMap, and Strike.
Schrödinger Release - New Features 2021-3
Schrödinger, LLC provides scientific software solutions and services for life sciences and materials research, as well as academic, government, and non-profit institutions around the world. It offers small-molecule drug discovery, biologics, materials science, and discovery informatics solutions; and PyMOL, a 3D molecular visualization solution. The company was founded in 1990 and is based in Portland, Oregon with operations in the United States, Europe, Japan, and India.
Product: Schrödinger Suites
Version: 2021-3 Build 123
Supported Architectures: x64
Website Home Page : www.schrodinger.com
Languages Supported: english
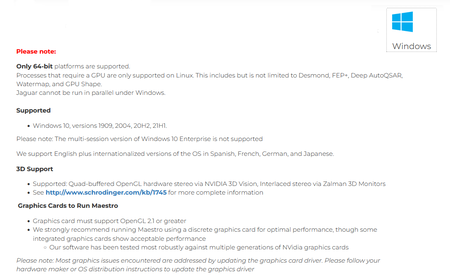
System Requirements: Windows *
Size: 6.5 Gb
Please visit my blog
Added by 3% of the overall size of the archive of information for the restoration
No mirrors please
Added by 3% of the overall size of the archive of information for the restoration
No mirrors please